Metabolismo: diferenças entre revisões
m |
+ trad |
||
| Linha 1: | Linha 1: | ||
{{tradução}} |
|||
[[Image:ATP-3D-vdW.png|thumb|right|280px|Estrutura do [[trifosfato de adenosina]], um intermediário central no metabolismo energético.]] |
[[Image:ATP-3D-vdW.png|thumb|right|280px|Estrutura do [[trifosfato de adenosina]], um intermediário central no metabolismo energético.]] |
||
'''Metabolismo''' (do [[língua grega|grego]] ''metabolé'', μεταβολισμος, que significa "mudança", troca, acrescido de ''ismo'') é o conjunto de [[reacção química|transformações]] que as [[substância]]s [[química]]s sofrem no interior dos [[organismo]]s vivos. É também usado o termo "'''metabolismo celular'''" referindo-se mais especificamente ao conjunto de todas as reacções químicas que ocorrem nas [[célula]]s. Estas [[reação química|reações químicas]] são responsáveis pelos processos de [[síntese]] e degradação dos [[nutriente]]s na [[célula]]. |
'''Metabolismo''' (do [[língua grega|grego]] ''metabolé'', μεταβολισμος, que significa "mudança", troca, acrescido de ''ismo'') é o conjunto de [[reacção química|transformações]] que as [[substância]]s [[química]]s sofrem no interior dos [[organismo]]s vivos. É também usado o termo "'''metabolismo celular'''" referindo-se mais especificamente ao conjunto de todas as reacções químicas que ocorrem nas [[célula]]s. Estas [[reação química|reações químicas]] são responsáveis pelos processos de [[síntese]] e degradação dos [[nutriente]]s na [[célula]]. |
||
| Linha 6: | Linha 7: | ||
As reacções químicas do metabolismo estão organizadas em [[via metabólica|vias metabólicas]], que são sequências de reacções em que o [[produto]] de uma reacção é utilizado como [[reagente]] na reacção seguinte. Diferentes [[enzima]]s [[catálise enzimática|catalisam]] diferentes passos de vias metabólicas, agindo de forma concertada de modo a não interromper o fluxo nessas vias. As enzimas são vitais para o metabolismo porque permitem a realização de reacções desejáveis mas [[termodinâmica|termodinamicamente]] desfavoráveis, ao acoplá-las a reacções mais favoráveis. As enzimas [[Regulação da atividade das enzimas|regulam]] as vias metabólicas em resposta a mudanças no ambiente celular ou a [[sinalização celular|sinais]] de outras células. |
As reacções químicas do metabolismo estão organizadas em [[via metabólica|vias metabólicas]], que são sequências de reacções em que o [[produto]] de uma reacção é utilizado como [[reagente]] na reacção seguinte. Diferentes [[enzima]]s [[catálise enzimática|catalisam]] diferentes passos de vias metabólicas, agindo de forma concertada de modo a não interromper o fluxo nessas vias. As enzimas são vitais para o metabolismo porque permitem a realização de reacções desejáveis mas [[termodinâmica|termodinamicamente]] desfavoráveis, ao acoplá-las a reacções mais favoráveis. As enzimas [[Regulação da atividade das enzimas|regulam]] as vias metabólicas em resposta a mudanças no ambiente celular ou a [[sinalização celular|sinais]] de outras células. |
||
O metabolismo de um organismo determina que substâncias são [[nutriente|nutricionais]] e quais são [[toxina|tóxicas]]. Por exemplo, alguns [[procariontes]] utilizam [[ácido sulfídrico]] como nutriente; este gás é no entanto [[veneno]]so para [[animais]]<ref>{{cite journal |author=Friedrich C |title=Physiology and genetics of sulfur-oxidizing bacteria |journal=Adv Microb Physiol |volume=39 |issue= |pages=235-89 |year=1998 |pmid=9328649}}</ref>. A [[velocidade]] a que se processa o metabolismo, determinada pela taxa metabólica, também influencia a quantidade de [[alimento]] requerida por um organismo. |
|||
Uma característica do metabolismo é a semelhança de vias metabólicas básicas entre [[espécie]]s muito diferentes. Por exemplo, o conjunto de intermediários reaccionais encontrados no [[ciclo dos ácidos tricarboxílicos]] é encontrado de forma universal, em células tão diferentes como a [[bactéria]] ''[[Escherichia coli]]'' ou o [[elefante]]<ref name=SmithE>{{cite journal |author=Smith E, Morowitz H |title=Universality in intermediary metabolism |url=http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=15340153 |journal=Proc Natl Acad Sci U S A |volume=101 |issue=36 |pages=13168-73 |year=2004 |pmid=15340153}}</ref>. Esta estrutura metabólica semelhante está provavelmente associada à grande eficiência dessas vias e na sua antiguidade na [[evolução|história da evolução]]<ref name=Ebenhoh>{{cite journal |author=Ebenhöh O, Heinrich R |title=Evolutionary optimization of metabolic pathways. Theoretical reconstruction of the stoichiometry of ATP and NADH producing systems |journal=Bull Math Biol |volume=63 |issue=1 |pages=21–55 |year=2001 |pmid=11146883}}</ref><ref name=Cascante>{{cite journal |author=Meléndez-Hevia E, Waddell T, Cascante M |title=The puzzle of the Krebs citric acid cycle: assembling the pieces of chemically feasible reactions, and opportunism in the design of metabolic pathways during evolution |journal=J Mol Evol |volume=43 |issue=3 |pages=293–303 |year=1996 |pmid=8703096}}</ref>. |
|||
O metabolismo é fundamentalmente estudado pela [[Bioquímica]], usando muitas vezes também técnicas ligadas à [[Biologia Molecular]] e à [[Genética]]. |
|||
==Substâncias bioquímicas relevantes== |
|||
[[Image:Trimyristin-3D-vdW.png|right|thumb|250px|Estrutura de um [[triacilglicerol]].]] |
|||
A maioria das estruturas que compõem os seres vivos é fabricada a partir de três classes básicas de moléculas: [[aminoácido]]s, [[glícido]]s e [[lípido]]s. Como estas moléculas são vitais, o metabolismo concentra-se no fabrico destas, na construção de células e [[tecido]]s ou na sua degradação para uso como fonte de [[energia]]. Muitos compostos bioquímicos podem ser condensados formando [[polímero]]s, como o [[ADN]] e as [[proteína]]s. Estas [[macromolécula]]s são parte essencial de todos os organismos vivos. |
|||
Alguns dos polímeros mais comuns estão listados abaixo: |
|||
{| class="wikitable" style="margin-left: auto; margin-right: auto;" |
|||
!Tipo de molécula |
|||
!Nome da forma monomérica |
|||
!Nome da forma polimérica |
|||
!Exemplos de formas poliméricas |
|||
|- |
|||
|align="center" |[[Aminoácido]]s |
|||
|align="center" |Aminoácidos |
|||
|align="center" |[[Proteína]]s (ou [[polipéptido]]s) |
|||
|align="center" |[[Proteínas fibrilares]] e [[proteínas globulares]] |
|||
|- |
|||
|align="center" |[[Glícido]]s |
|||
|align="center" |[[Monossacarídeo]]s |
|||
|align="center" |[[Polissacarídeo]]s |
|||
|align="center" |[[Amido]], [[glicogénio]] e [[celulose]] |
|||
|- |
|||
|align="center" |[[Ácido nucleico|Ácidos nucleicos]] |
|||
|align="center" |[[Nucleótidos]] |
|||
|align="center" |[[Polinucleótido]]s |
|||
|align="center" |[[ADN]] e [[ARN]] |
|||
|} |
|||
===Aminoácidos e proteínas=== |
|||
As [[proteínas]] são compostas por aminoácidos dispostos numa cadeia linear e ligados entre si por [[ligação peptídica|ligações peptídicas]]. Muitas proteínas são as enzimas que catalisam as reacções químicas no metabolismo. Outras proteínas têm funções estruturais ou mecânicas, como o sistema de armação celular usado para manter a forma da célula, o [[citoesqueleto]]<ref>{{cite journal |author=Michie K, Löwe J |title=Dynamic filaments of the bacterial cytoskeleton |journal=Annu Rev Biochem |volume=75 |issue= |pages=467-92 |year=2006 |pmid=16756499}}</ref>. |
|||
As proteínas desempenham também papéis importantes na [[sinalização celular]], [[imunoglobulina|resposta imunitária]], [[adesão celular]], [[transporte activo]] através de [[membrana]]s e no [[ciclo celular]]<ref name=Nelson>{{cite book | last = Nelson | first = David L. | coauthors = Michael M. Cox | title = Lehninger Principles of Biochemistry | publisher = W. H. Freeman and company | date = 2005 | location = New York | pages = 841 | isbn = 0-7167-4339-6}}</ref>. |
|||
===Lípidos=== |
|||
Os [[lípidos]] são o grupo mais diversificado de compostos bioquímicos. Constituem grande parte das membranas biológicas, tais como a [[membrana celular]]; além desta função estrutural, também servem como fonte de energia<ref name=Nelson/>. Os lípidos são normalmente definidos como moléculas biológicas [[hidrofóbica]]s ou [[anfipática]]s [[solubilidade|solúveis]] em [[solvente]]s [[composto orgânico|orgânicos]] como o [[benzeno]] ou o [[clorofórmio]]<ref>{{cite journal |author=Fahy E, Subramaniam S, Brown H, Glass C, Merrill A, Murphy R, Raetz C, Russell D, Seyama Y, Shaw W, Shimizu T, Spener F, van Meer G, VanNieuwenhze M, White S, Witztum J, Dennis E |title=A comprehensive classification system for lipids |url=http://www.jlr.org/cgi/content/full/46/5/839 |journal=J Lipid Res |volume=46 |issue=5 |pages=839-61 |year=2005 |pmid=15722563}}</ref>. |
|||
As [[gordura]]s são um grupo alargado de compostos que inclui os [[ácidos gordos]] e o [[glicerol]]; uma molécula de glicerol ligada a três ácidos gordos por uma ligação [[éster]] é um [[triacilglicerol]]<ref>{{cite web | title=Nomenclature of Lipids |publisher=IUPAC-IUB Commission on Biochemical Nomenclature (CBN) | url=http://www.chem.qmul.ac.uk/iupac/lipid/ |accessdate=2007-03-08}}</ref>. Existem diversas variações desta estrutura básica, incluindo a presença de [[esfingosina]] em [[esfingolípido]]s e grupos hidrofílicos como o [[fosfato]] nos [[fosfolípido]]s. |
|||
Os [[esteróide]]s, como o [[colesterol]], são outro grupo significativo de lípidos sintetizados em células.<ref>{{cite journal |author=Hegardt F |title=Mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase: a control enzyme in ketogenesis |url=http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=1220089&blobtype=pdf |journal=Biochem J |volume=338 (Pt 3) |issue= |pages=569-82 |year=1999 |pmid=10051425}}</ref> |
|||
===Glícidos=== |
|||
[[Image:Glucose Fisher to Haworth.gif|thumb|230px|right|Estrutura da [[glicose]] convertida da projecção de Fisher para a de Haworth.]] |
|||
Os [[glícidos]] são [[aldeído]]s ou [[cetona]]s contendo diversos [[grupo funcional|grupos funcionais]] [[hidroxilo]]. Os glícidos simples podem existir numa forma linear ou numa forma cíclica. São as moléculas biológicas mais abundantes e possuem funções muito diversificadas, como o armazenamento e transporte de energia (sob a forma de [[amido]] e [[glicogénio]]) e construção de elementos estruturais (como a [[celulose]] em [[plantas]] e a [[quitina]] em [[animais]])<ref name=Nelson/>. |
|||
Os glícidos mais simples são os [[monossacarídeos]], que incluem a [[galactose]], a [[frutose]] e a [[glicose]]. Os monossacarídeos podem formar polímeros designados [[polissacarídeos]] de formas muito diversas<ref>{{cite journal |author=Raman R, Raguram S, Venkataraman G, Paulson J, Sasisekharan R |title=Glycomics: an integrated systems approach to structure-function relationships of glycans |journal=Nat Methods |volume=2 |issue=11 |pages=817-24 |year=2005 |pmid=16278650}}</ref>. |
|||
===Nucleótidos=== |
|||
Os polímeros [[ADN]] e [[ARN]] são longas cadeias de [[nucleótido]]s. Estas macromoléculas são essenciais no armazenamento e uso da [[código genético|informação genética]], através dos processos de [[transcrição]] e [[síntese proteica]]<ref name=Nelson/>. Esta informação é protegida por mecanismos de [[reparação do ADN]] e propagada através da [[replicação]] do ADN. Alguns [[vírus]] têm um genoma constituído por ARN (por exemplo, o [[HIV]]), que usam [[transcrição reversa]] para sintetizar ADN a partir desse ARN<ref>{{cite journal |author=Sierra S, Kupfer B, Kaiser R |title=Basics of the virology of HIV-1 and its replication |journal=J Clin Virol |volume=34 |issue=4 |pages=233-44 |year=2005 |pmid=16198625}}</ref>. |
|||
O ARN de [[ribozima]]s (como o [[spliceossoma]]) apresenta actividade enzimática tal como as enzimas proteicas, pois pode catalisar reacções químicas. |
|||
Os [[nucleósido]]s são sintetizados a partir da [[ligação química|ligação]] de uma [[base azotada]] a uma [[ribose]]. Estas bases são [[Composto heterocíclico|anéis heterocíclicos]] contendo [[azoto]], classificados como [[purina]]s ou [[pirimidina]]s. Os nucleótidos também actuam como [[coenzima]]s em reacções de transferência de grupos químicos<ref name=Wimmer>{{cite journal |author=Wimmer M, Rose I |title=Mechanisms of enzyme-catalyzed group transfer reactions |journal=Annu Rev Biochem |volume=47 |issue= |pages=1031–78 |year=1978 |pmid=354490}}</ref>. |
|||
<!-- ===Coenzymes=== |
|||
[[Image:Acetyl-CoA-2D.svg|thumb|right|300px|Structure of the [[coenzyme]] [[acetyl-CoA]].The transferable [[acetyl|acetyl group]] is bonded to the sulphur atom at the extreme left.]] |
|||
Metabolism involves a vast array of chemical reactions, but most fall under a few basic types of [[functional group|group transfer reaction]]s.<ref>{{cite journal |author=Mitchell P |title=The Ninth Sir Hans Krebs Lecture. Compartmentation and communication in living systems. Ligand conduction: a general catalytic principle in chemical, osmotic and chemiosmotic reaction systems |journal=Eur J Biochem |volume=95 |issue=1 |pages=1–20 |year=1979 |pmid=378655}}</ref> This common chemistry allows cells to use a small set of metabolic intermediates to carry chemical groups between different reactions.<ref name=Wimmer/> These group-transfer intermediates are called [[coenzyme]]s. Each class of group-transfer reaction is carried out by a particular coenzyme, which is the [[substrate (biochemistry)|substrate]] for a set of enzymes that produce it, and a set of enzymes that consume it. These coenzymes are therefore continuously being made, consumed and then recycled.<ref name=Dimroth>{{cite journal |author=Dimroth P, von Ballmoos C, Meier T |title=Catalytic and mechanical cycles in F-ATP synthases. Fourth in the Cycles Review Series |url=http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=16607397 |journal=EMBO Rep |volume=7 |issue=3 |pages=276-82 |year=2006 |pmid=16607397}}</ref> |
|||
The most central coenzyme is [[adenosine triphosphate]] (ATP), the universal energy currency of cells. This [[nucleotide]] is used to transfer chemical energy between different chemical reactions. There is only a small amount of ATP in cells, but as it is continuously regenerated, the human body can use about its own weight in ATP per day.<ref name=Dimroth/> ATP acts as a bridge between catabolism and anabolism, with catabolic reactions generating ATP and anabolic reactions consuming it. It also serves as a carrier of phosphate groups in [[phosphorylation]] reactions. |
|||
A [[vitamin]] is an organic compound needed in small quantities that cannot be made in the cells. In human [[nutrition]], most vitamins function as coenzymes after modification; for example, all water-soluble vitamins are phosphorylated or are coupled to nucleotides when they are used in cells.<ref>{{Citation | last1 = Coulston | first1 = Ann | last2 = Kerner | first2 = John | last3 = Hattner | first3 = JoAnn | last4 = Srivastava | first4 = Ashini | contribution = Nutrition Principles and Clinical Nutrition | title = Stanford School of Medicine Nutrition Courses | publisher = SUMMIT | year = 2006 }}</ref> [[Nicotinamide adenine dinucleotide]] (NADH), a derivative of vitamin B<sub>3</sub> ([[niacin]]), is an important coenzyme that acts as a hydrogen acceptor. Hundreds of separate types of [[dehydrogenase]]s remove electrons from their substrates and [[redox|reduce]] NAD<sup>+</sup> into NADH. This reduced form of the coenzyme is then a substrate for any of the [[reductase]]s in the cell that need to reduce their substrates.<ref>{{cite journal |author=Pollak N, Dölle C, Ziegler M |title=The power to reduce: pyridine nucleotides—small molecules with a multitude of functions |journal=Biochem J |volume=402 |issue=2 |pages=205-18 |year=2007 |pmid=17295611}}</ref> Nicotinamide adenine dinucleotide exists in two related forms in the cell, NADH and NADPH. The NAD<sup>+</sup>/NADH form is more important in catabolic reactions, while NADP<sup>+</sup>/NADPH is used in anabolic reactions. |
|||
[[Image:1GZX Haemoglobin.png|thumb|300px|left|Structure of [[hemoglobin]]. The protein subunits are in red and blue, and the iron-containing [[heme]] groups in green. From {{PDB|1GZX}}.]] |
|||
===Minerals and cofactors=== |
|||
{{further|[[Physiology]], [[bioinorganic chemistry]] and [[Human iron metabolism|iron metabolism]]}} |
|||
Inorganic elements play critical roles in metabolism; some are abundant (e.g. [[sodium]] and [[potassium]]) while others function at minute concentrations. About 99% of mammals' mass are the elements [[carbon]], [[nitrogen]], [[calcium]], [[sodium]], [[chlorine]], [[potassium]], [[hydrogen]], [[oxygen]] and [[sulfur]].<ref name=Heymsfield>{{cite journal |author=Heymsfield S, Waki M, Kehayias J, Lichtman S, Dilmanian F, Kamen Y, Wang J, Pierson R |title=Chemical and elemental analysis of humans in vivo using improved body composition models |journal=Am J Physiol |volume=261 |issue=2 Pt 1 |pages=E190-8 |year=1991 |pmid=1872381}}</ref> The [[organic compound]]s (proteins, lipids and carbohydrates) contain the majority of the carbon and nitrogen and most of the oxygen and hydrogen is present as water.<ref name=Heymsfield/> |
|||
The abundant inorganic elements act as [[ion]]ic [[electrolyte]]s. The most important ions are [[sodium]], [[potassium]], [[calcium]], [[magnesium]], [[chloride]], [[phosphate]], and the organic ion [[bicarbonate]]. The maintenance of precise [[ion gradient|gradient]]s across cell membranes maintains [[osmotic pressure]] and [[pH]].<ref>{{cite journal |author=Sychrová H |title=Yeast as a model organism to study transport and homeostasis of alkali metal cations |url=http://www.biomed.cas.cz/physiolres/pdf/53%20Suppl%201/53_S91.pdf |journal=Physiol Res |volume=53 Suppl 1 |issue= |pages=S91-8 |year=2004 |pmid=15119939}}</ref> Ions are also critical for [[nerve]]s and [[muscle]]s, as [[action potential]]s in these tissues are produced by the exchange of electrolytes between the [[extracellular fluid]] and the [[cytosol]].<ref>{{cite journal |author=Levitan I |title=Modulation of ion channels in neurons and other cells |journal=Annu Rev Neurosci |volume=11 |issue= |pages=119-36 |year=1988 |pmid=2452594}}</ref> Electrolytes enter and leave cells through proteins in the [[cell membrane]] called [[ion channels]]. For example, [[muscle contraction]] depends upon the movement of calcium, sodium and potassium through ion channels in the cell membrane and [[T-tubule]]s.<ref>{{cite journal |author=Dulhunty A |title=Excitation-contraction coupling from the 1950s into the new millennium |journal=Clin Exp Pharmacol Physiol |volume=33 |issue=9 |pages=763-72 |year=2006 |pmid=16922804}}</ref> |
|||
The [[transition metal]]s are usually present as [[trace element]]s in organisms, with [[zinc]] and [[iron]] being most abundant.<ref>{{cite journal |author=Mahan D, Shields R |title=Macro- and micromineral composition of pigs from birth to 145 kilograms of body weight |url=http://jas.fass.org/cgi/reprint/76/2/506 |journal=J Anim Sci |volume=76 |issue=2 |pages=506-12 |year=1998 |pmid=9498359}}</ref><ref name=Husted>{{cite journal |author=Husted S, Mikkelsen B, Jensen J, Nielsen N |title=Elemental fingerprint analysis of barley (Hordeum vulgare) using inductively coupled plasma mass spectrometry, isotope-ratio mass spectrometry, and multivariate statistics |journal=Anal Bioanal Chem |volume=378 |issue=1 |pages=171-82 |year=2004 |pmid=14551660}}</ref> These metals are used in some proteins as cofactors and are essential for the activity of enzymes such as [[catalase]] and oxygen-carrier proteins such as [[hemoglobin]].<ref>{{cite journal |author=Finney L, O'Halloran T |title=Transition metal speciation in the cell: insights from the chemistry of metal ion receptors |journal=Science |volume=300 |issue=5621 |pages=931-6 |year=2003 |pmid=12738850}}</ref> These [[cofactors]] are bound tightly to a specific protein; although enzyme cofactors can be modified during catalysis, cofactors always return to their original state after catalysis has taken place. The metal micronutrients are taken up into organisms by specific transporters and bound to storage proteins such as [[ferritin]] or [[metallothionein]] when not being used.<ref>{{cite journal |author=Cousins R, Liuzzi J, Lichten L |title=Mammalian zinc transport, trafficking, and signals |url=http://www.jbc.org/cgi/content/full/281/34/24085 |journal=J Biol Chem |volume=281 |issue=34 |pages=24085-9 |year=2006 |pmid=16793761}}</ref><ref>{{cite journal |author=Dunn L, Rahmanto Y, Richardson D |title=Iron uptake and metabolism in the new millennium |journal=Trends Cell Biol |volume=17 |issue=2 |pages=93–100 |year=2007 |pmid=17194590}}</ref> |
|||
== Catabolism == |
|||
{{further|[[Catabolism]]}} |
|||
Catabolism is the set of metabolic processes that release energy. These include breaking down and oxidising food molecules as well as reactions that trap the energy in sunlight. The purpose of these catabolic reactions is to provide the energy and components needed by anabolic reactions. The exact nature of these catabolic reactions differ from organism to organism, with organic molecules being used as a source of energy in [[organotroph]]s, while [[lithotroph]]s use inorganic substrates and [[phototroph]]s capture [[sunlight]] as [[potential energy#Chemical energy|chemical energy]]. However, all these different forms of metabolism depend on [[redox]] reactions that involve the transfer of electrons from reduced donor molecules such as [[organic molecule]]s, [[water]], [[ammonia]], [[hydrogen sulfide]] or [[Ferrous|ferrous ions]] to acceptor molecules such as [[oxygen]], [[nitrate]] or [[sulphate]].<ref>{{cite journal |author=Nealson K, Conrad P |title=Life: past, present and future |url=http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=10670014 |journal=Philos Trans R Soc Lond B Biol Sci |volume=354 |issue=1392 |pages=1923–39 |year=1999 |pmid=10670014}}</ref> In animals these reactions involve complex [[organic molecule]]s being broken down to simpler molecules, such as [[carbon dioxide]] and water. In [[photosynthesis|photosynthetic]] organisms such as plants and [[cyanobacteria]], these electron-transfer reactions do not release energy, but are used as a way of storing energy absorbed from sunlight.<ref name=Nelson>{{cite journal |author=Nelson N, Ben-Shem A |title=The complex architecture of oxygenic photosynthesis |journal=Nat Rev Mol Cell Biol |volume=5 |issue=12 |pages=971-82 |year=2004 |pmid=15573135}}</ref> |
|||
The most common set of catabolic reactions in animals can be separated into three main stages. In the first, large organic molecules such as [[protein]]s, [[polysaccharide]]s or [[lipid]]s are digested into their smaller components outside cells. Next, these smaller molecules are taken up by cells and converted to yet smaller molecules, usually [[coenzyme A|acetyl coenzyme A]], which releases some energy. Finally, the acetyl group on the coenzyme A is oxidised to water and carbon dioxide, releasing energy that is stored by reducing the coenzyme [[nicotinamide adenine dinucleotide]] (NAD<sup>+</sup>) into NADH. |
|||
===Digestion=== |
|||
{{further|[[Digestion]] and [[gastrointestinal tract]]}} |
|||
Macromolecules such as starch, cellulose or proteins cannot be rapidly taken up by cells and need to be broken into their smaller units before they can be used in cell metabolism. Several common classes of enzymes digest these polymers. These digestive enzymes include [[protease]]s that digest proteins into amino acids, as well as [[glycoside hydrolase]]s that digest polysaccharides into monosaccharides. |
|||
Microbes simply secrete digestive enzymes into their surroundings,<ref>{{cite journal |author=Häse C, Finkelstein R |title=Bacterial extracellular zinc-containing metalloproteases |url=http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=8302217 |journal=Microbiol Rev |volume=57 |issue=4 |pages=823-37 |year=1993 |pmid=8302217}}</ref><ref>{{cite journal |author=Gupta R, Gupta N, Rathi P |title=Bacterial lipases: an overview of production, purification and biochemical properties |journal=Appl Microbiol Biotechnol |volume=64 |issue=6 |pages=763-81 |year=2004 |pmid=14966663}}</ref> while animals only secrete these enzymes from specialized cells in their [[gut]]s.<ref>{{cite journal |author=Hoyle T |title=The digestive system: linking theory and practice |journal=Br J Nurs |volume=6 |issue=22 |pages=1285–91 |year=1997 |pmid=9470654}}</ref> The amino acids or sugars released by these extracellular enzymes are then pumped into cells by specific [[active transport]] proteins.<ref>{{cite journal |author=Souba W, Pacitti A |title=How amino acids get into cells: mechanisms, models, menus, and mediators |journal=JPEN J Parenter Enteral Nutr |volume=16 |issue=6 |pages=569-78 |year=1992 |pmid=1494216}}</ref><ref>{{cite journal |author=Barrett M, Walmsley A, Gould G |title=Structure and function of facilitative sugar transporters |journal=Curr Opin Cell Biol |volume=11 |issue=4 |pages=496–502 |year=1999 |pmid=10449337}}</ref> |
|||
[[Image:Catabolism schematic.svg|thumb|left|300px|A simplified outline of the catabolism of [[protein]]s, [[carbohydrate]]s and [[fat]]s.]] |
|||
===Energy from organic compounds=== |
|||
{{further|[[Cellular respiration]], [[Fermentation (biochemistry)|fermentation]], [[carbohydrate catabolism]], [[fat catabolism]] and [[protein catabolism]]}} |
|||
Carbohydrate catabolism is the breakdown of carbohydrates into smaller units. Carbohydrates are usually taken into cells once they have been digested into [[monosaccharide]]s.<ref>{{cite journal |author=Bell G, Burant C, Takeda J, Gould G |title=Structure and function of mammalian facilitative sugar transporters |journal=J Biol Chem |volume=268 |issue=26 |pages=19161-4 |year=1993 |pmid=8366068}}</ref> Once inside, the major route of breakdown is [[glycolysis]], where sugars such as [[glucose]] and [[fructose]] are converted into [[pyruvic acid|pyruvate]] and some ATP is generated.<ref name=Bouche>{{cite journal |author=Bouché C, Serdy S, Kahn C, Goldfine A |title=The cellular fate of glucose and its relevance in type 2 diabetes |url=http://edrv.endojournals.org/cgi/content/full/25/5/807 |journal=Endocr Rev |volume=25 |issue=5 |pages=807-30 |year=2004 |pmid=15466941}}</ref> Pyruvate is an intermediate in several metabolic pathways, but the majority is converted to [[acetyl-CoA]] and fed into the [[citric acid cycle]]. Although some more ATP is generated in the citric acid cycle, the most important product is NADH, which is made from NAD<sup>+</sup> as the acetyl-CoA is oxidized. This oxidation releases [[carbon dioxide]] as a waste product. An alternative route for glucose breakdown is the [[pentose phosphate pathway]], which reduces the coenzyme [[NADPH]] and produces [[pentose]] sugars such as [[ribose]], the sugar component of [[nucleic acid]]s. |
|||
Fats are catabolised by [[hydrolysis]] to free fatty acids and glycerol. The glycerol enters glycolysis and the fatty acids are broken down by [[beta oxidation]] to release acetyl-CoA, which then is fed into the citric acid cycle. Due to their high proportion of [[methylene]] groups, fatty acids release more energy upon oxidation than carbohydrates, as carbohydrates such as glucose contain more oxygen in their structures. |
|||
[[Amino acid]]s are either used to synthesize proteins and other biomolecules, or oxidized to [[urea]] and carbon dioxide as a source of energy.<ref>{{cite journal |author=Sakami W, Harrington H |title=Amino acid metabolism |journal=Annu Rev Biochem |volume=32 |issue= |pages=355-98 |year=1963 |pmid=14144484}}</ref> The oxidation pathway starts with the removal of the amino group by a [[transaminase]]. The amino group is fed into the [[urea cycle]], leaving a deaminated carbon skeleton in the form of a [[keto acid]]. Several of these keto acids are intermediates in the citric acid cycle, for example the deamination of [[glutamate]] forms α-[[Ketoglutaric acid|ketoglutarate]].<ref>{{cite journal |author=Brosnan J |title=Glutamate, at the interface between amino acid and carbohydrate metabolism |url=http://jn.nutrition.org/cgi/content/full/130/4/988S |journal=J Nutr |volume=130 |issue=4S Suppl |pages=988S-90S |year=2000 |pmid=10736367}}</ref> The [[glucogenic amino acid]]s can also be converted into glucose, through [[gluconeogenesis]] (discussed below).<ref>{{cite journal |author=Young V, Ajami A |title=Glutamine: the emperor or his clothes? |url=http://jn.nutrition.org/cgi/content/full/131/9/2449S |journal=J Nutr |volume=131 |issue=9 Suppl |pages=2449S-59S; discussion 2486S-7S |year=2001 |pmid=11533293}}</ref> |
|||
===Oxidative phosphorylation=== |
|||
[[image:Atp_synthase.PNG|right|thumb|280px|Structure of [[ATP synthase]], the proton channel is shown in blue and the synthase subunit in red.]] |
|||
{{further|[[Oxidative phosphorylation]], [[chemiosmosis]] and [[mitochondrion]]}} |
|||
In oxidative phosphorylation, the electrons removed from food molecules in pathways such as the citric acid cycle are transferred to oxygen and the energy released used to make ATP. This is done in [[eukaryote]]s by a series of proteins in the membranes of mitochondria called the [[electron transport chain]]. In [[prokaryote]]s, these proteins are found in the cell's [[bacterial cell structure|inner membrane]].<ref>{{cite journal |author=Hosler J, Ferguson-Miller S, Mills D |title=Energy transduction: proton transfer through the respiratory complexes |journal=Annu Rev Biochem |volume=75 |issue= |pages=165-87 |year=2006 |pmid=16756489}}</ref> These proteins use the energy released from oxidising the electron-carrying coenzyme NADH to pump [[proton]]s across the membrane.<ref>{{cite journal |author=Schultz B, Chan S |title=Structures and proton-pumping strategies of mitochondrial respiratory enzymes |journal=Annu Rev Biophys Biomol Struct |volume=30 |issue= |pages=23–65 |year=2001 |pmid=11340051}}</ref> |
|||
Pumping protons out of the mitochondria creates a proton [[diffusion|concentration difference]] across the membrane and generates a [[electrochemical gradient]].<ref>{{cite journal |author=Capaldi R, Aggeler R |title=Mechanism of the F(1)F(0)-type ATP synthase, a biological rotary motor |journal=Trends Biochem Sci |volume=27 |issue=3 |pages=154-60 |year=2002 |pmid=11893513}}</ref> This force drives protons back into the mitochondrion through the stalk subunit of the [[ATP synthase]]. The flow of protons makes the lower subunit rotate, causing its [[active site]] to phosphorylate [[adenosine diphosphate]] and turn it into ATP.<ref name=Dimroth/> |
|||
===Energy from inorganic compounds=== |
|||
{{further|[[Microbial metabolism]] and [[nitrogen cycle]]}} |
|||
[[Chemolithotroph]]y is a type of metabolism found in [[prokaryote]]s where energy is obtained from the oxidation of [[inorganic compounds]]. These organisms can use [[hydrogen]],<ref>{{cite journal |author=Friedrich B, Schwartz E |title=Molecular biology of hydrogen utilization in aerobic chemolithotrophs |journal=Annu Rev Microbiol |volume=47 |issue= |pages=351-83 |year=1993 |pmid=8257102}}</ref> reduced [[sulfur]] compounds (such as [[sulfide]], [[hydrogen sulfide]] and [[thiosulfate]]),<ref>{{cite journal |author=Friedrich C |title=Physiology and genetics of sulfur-oxidizing bacteria |journal=Adv Microb Physiol |volume=39 |issue= |pages=235-89 |year=1998 |pmid=9328649}}</ref> [[Iron(II) oxide|ferrous iron (FeII)]]<ref>{{cite journal |author=Weber K, Achenbach L, Coates J |title=Microorganisms pumping iron: anaerobic microbial iron oxidation and reduction |journal=Nat Rev Microbiol |volume=4 |issue=10 |pages=752-64 |year=2006 |pmid=16980937}}</ref> or [[ammonia]]<ref>{{cite journal |author=Jetten M, Strous M, van de Pas-Schoonen K, Schalk J, van Dongen U, van de Graaf A, Logemann S, Muyzer G, van Loosdrecht M, Kuenen J |title=The anaerobic oxidation of ammonium |journal=FEMS Microbiol Rev |volume=22 |issue=5 |pages=421-37 |year=1998 |pmid=9990725}}</ref> as sources of reducing power and they gain energy from the oxidation of these compounds with electron acceptors such as [[oxygen]] or [[nitrite]].<ref>{{cite journal |author=Simon J |title=Enzymology and bioenergetics of respiratory nitrite ammonification |journal=FEMS Microbiol Rev |volume=26 |issue=3 |pages=285–309 |year=2002 |pmid=12165429}}</ref> These microbial processes are important in global [[biogeochemical cycle]]s such as [[acetogenesis]], [[nitrification]] and [[denitrification]] and are critical for [[fertility (soil)|soil fertility]].<ref>{{cite journal |author=Conrad R |title=Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO) |url=http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=8987358 |journal=Microbiol Rev |volume=60 |issue=4 |pages=609-40 |year=1996 |pmid=8987358}}</ref><ref>{{cite journal |author=Barea J, Pozo M, Azcón R, Azcón-Aguilar C |title=Microbial co-operation in the rhizosphere |url=http://jxb.oxfordjournals.org/cgi/content/full/56/417/1761 |journal=J Exp Bot |volume=56 |issue=417 |pages=1761–78 |year=2005 |pmid=15911555}}</ref> |
|||
===Energy from light=== |
|||
{{further|[[Phototroph]], [[photophosphorylation]], [[chloroplast]]}} |
|||
The energy in sunlight is captured by [[plants]], [[cyanobacteria]], [[purple bacteria]], [[green sulfur bacteria]] and some [[protist]]s. This process is often coupled to the conversion of carbon dioxide into organic compounds, as part of photosynthesis, which is discussed below. The energy capture and carbon fixation systems can however operate separately in prokaryotes, as purple bacteria and green sulfur bacteria can use sunlight as a source of energy, while switching between carbon fixation and the fermentation of organic compounds.<ref>{{cite journal |author=van der Meer M, Schouten S, Bateson M, Nübel U, Wieland A, Kühl M, de Leeuw J, Sinninghe Damsté J, Ward D |title=Diel variations in carbon metabolism by green nonsulfur-like bacteria in alkaline siliceous hot spring microbial mats from Yellowstone National Park |url=http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=16000812 |journal=Appl Environ Microbiol |volume=71 |issue=7 |pages=3978–86 |year=2005 |pmid=16000812}}</ref><ref>{{cite journal |author=Tichi M, Tabita F |title=Interactive control of Rhodobacter capsulatus redox-balancing systems during phototrophic metabolism |url=http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=11591679 |journal=J Bacteriol |volume=183 |issue=21 |pages=6344–54 |year=2001 |pmid=11591679}}</ref> |
|||
The capture of solar energy is a process that is similar in principle to oxidative phosphorylation, as it involves energy being stored as a proton concentration gradient and this proton motive force then driving ATP synthesis.<ref name=Dimroth/> The electrons needed to drive this electron transport chain come from light-gathering proteins called [[photosynthetic reaction centre]]s. These structures are classed into two types depending on the type of [[photosynthetic pigment]] present, with most photosynthetic bacteria only having one type of reaction center, while plants and cyanobacteria have two.<ref>{{cite journal |author=Allen J, Williams J |title=Photosynthetic reaction centers |journal=FEBS Lett |volume=438 |issue=1–2 |pages=5–9 |year=1998 |pmid=9821949}}</ref> |
|||
In plants, [[photosystem|photosystem II]] uses light energy to remove electrons from water, releasing oxygen as a waste product. The electrons then flow to the [[cytochrome b6f complex]], which uses their energy to pump protons across the [[thylakoid]] membrane in the [[chloroplast]].<ref>{{cite journal |author=Nelson N, Ben-Shem A |title=The complex architecture of oxygenic photosynthesis |journal=Nat Rev Mol Cell Biol |volume=5 |issue=12 |pages=971-82 |year=2004 |pmid=15573135}}</ref> These protons move back through them membrane as they drive the ATP synthase, as before. The electrons then flow through [[photosystem|photosystem I]] and can then either be used to reduce the coenzyme NADP<sup>+</sup>, for use in the [[Calvin cycle]] which is discussed below, or recycled for further ATP generation.<ref>{{cite journal |author=Munekage Y, Hashimoto M, Miyake C, Tomizawa K, Endo T, Tasaka M, Shikanai T |title=Cyclic electron flow around photosystem I is essential for photosynthesis |journal=Nature |volume=429 |issue=6991 |pages=579-82 |year=2004 |pmid=15175756}}</ref> |
|||
== Anabolism == |
|||
{{further|[[Anabolism]]}} |
|||
'''Anabolism''' is the set of constructive metabolic processes where the energy released by catabolism is used to synthesize complex molecules. In general, the complex molecules that make up cellular structures are constructed step-by-step from small and simple precursors. Anabolism involves three basic stages. Firstly, the production of precursors such as [[amino acid]]s, [[monosaccharide]]s, [[Terpenoid|isoprenoids]] and [[nucleotide]]s, secondly, their activation into reactive forms using energy from ATP, and thirdly, the assembly of these precursors into complex molecules such as [[protein]]s, [[polysaccharide]]s, [[lipid]]s and [[nucleic acid]]s. |
|||
Organisms differ in how many of the molecules in their cells they can construct for themselves. [[Autotroph]]s such as plants can construct the complex organic molecules in cells such as polysaccharides and proteins from simple molecules like [[carbon dioxide]] and water. [[Heterotroph]]s, on the other hand, require a source of more complex substances, such as monosaccharides and amino acids, to produce these complex molecules. Organisms can be further classified by ultimate source of their energy: photoautotrophs and photoheterotrophs obtain energy from light, whereas chemoautotrophs and chemoheterotrophs obtain energy from inorganic oxidation reactions. |
|||
===Carbon fixation=== |
|||
{{further|[[Photosynthesis]], [[carbon fixation]] and [[chemosynthesis]]}} |
|||
[[Image:Chloroplasten.jpg|frame|Plant cells (bounded by purple walls) filled with chloroplasts (green), which are the site of photosynthesis.]] |
|||
Photosynthesis is the synthesis of glucose from sunlight, [[carbon dioxide]] (CO<sub>2</sub>) and water, with oxygen produced as a waste product. This process uses the ATP and NADPH produced by the [[photosynthetic reaction centre]]s, as described above, to convert CO<sub>2</sub> into [[glycerate 3-phosphate]], which can then be converted into glucose. This carbon-fixation reaction is carried out by the enzyme [[RuBisCO]] as part of the [[Calvin cycle|Calvin – Benson cycle]].<ref>{{cite journal |author=Miziorko H, Lorimer G |title=Ribulose-1,5-bisphosphate carboxylase-oxygenase |journal=Annu Rev Biochem |volume=52 |issue= |pages=507-35 |year=1983 |pmid=6351728}}</ref> Three types of photosynthesis occur in plants, [[C3 carbon fixation]], [[C4 carbon fixation]] and [[Crassulacean acid metabolism|CAM photosynthesis]]. These differ by the route that carbon dioxide takes to the Calvin cycle, with C3 plants fixing CO<sub>2</sub> directly, while C4 and CAM photosynthesis incorporate the CO<sub>2</sub> into other compounds first, as adaptations to deal with intense sunlight and dry conditions.<ref>{{cite journal |author=Dodd A, Borland A, Haslam R, Griffiths H, Maxwell K |title=Crassulacean acid metabolism: plastic, fantastic |url=http://jxb.oxfordjournals.org/cgi/content/full/53/369/569 |journal=J Exp Bot |volume=53 |issue=369 |pages=569-80 |year=2002 |pmid=11886877}}</ref> |
|||
In photosynthetic [[prokaryote]]s the mechanisms of carbon fixation are more diverse. Here, carbon dioxide can be fixed by the Calvin – Benson cycle, a [[Reverse Krebs cycle|reversed citric acid]] cycle,<ref>{{cite journal |author=Hügler M, Wirsen C, Fuchs G, Taylor C, Sievert S |title=Evidence for autotrophic CO2 fixation via the reductive tricarboxylic acid cycle by members of the epsilon subdivision of proteobacteria |url=http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=15838028 |journal=J Bacteriol |volume=187 |issue=9 |pages=3020–7 |year=2005 |pmid=15838028}}</ref> or the carboxylation of acetyl-CoA.<ref>{{cite journal |author=Strauss G, Fuchs G |title=Enzymes of a novel autotrophic CO2 fixation pathway in the phototrophic bacterium Chloroflexus aurantiacus, the 3-hydroxypropionate cycle |journal=Eur J Biochem |volume=215 |issue=3 |pages=633-43 |year=1993 |pmid=8354269}}</ref><ref>{{cite journal |author=Wood H |title=Life with CO or CO2 and H2 as a source of carbon and energy |url=http://www.fasebj.org/cgi/reprint/5/2/156 |journal=FASEB J |volume=5 |issue=2 |pages=156-63 |year=1991 |pmid=1900793}}</ref> Prokaryotic [[Chemotroph|chemoautotrophs]] also fix CO<sub>2</sub> through the Calvin – Benson cycle, but use energy from inorganic compounds to drive the reaction.<ref>{{cite journal |author=Shively J, van Keulen G, Meijer W |title=Something from almost nothing: carbon dioxide fixation in chemoautotrophs |journal=Annu Rev Microbiol |volume=52 |issue= |pages=191–230 |year=1998 |pmid=9891798}}</ref> |
|||
===Carbohydrates and glycans=== |
|||
{{further|[[Gluconeogenesis]], [[glyoxylate cycle]], [[glycogenesis]] and [[glycosylation]]}} |
|||
In carbohydrate anabolism, simple organic acids can be converted into [[monosaccharide]]s such as [[glucose]] and then used to assemble [[polysaccharide]]s such as [[starch]]. The generation of [[glucose]] from compounds like [[pyruvate]], [[lactate]], [[glycerol]], [[glycerate 3-phosphate]] and [[amino acids]] is called [[gluconeogenesis]]. Gluconeogenesis converts pyruvate to [[glucose-6-phosphate]] through a series of intermediates, many of which are shared with [[glycolysis]].<ref name=Bouche/> However, this pathway is not simply [[glycolysis]] run in reverse, as several steps are catalyzed by non-glycolytic enzymes. This is important as it allows the formation and breakdown of glucose to be regulated separately and prevents both pathways from running simultaneously in a [[futile cycle]].<ref>{{cite journal |author=Boiteux A, Hess B |title=Design of glycolysis |journal=Philos Trans R Soc Lond B Biol Sci |volume=293 |issue=1063 |pages=5–22 |year=1981 |pmid=6115423}}</ref><ref>{{cite journal |author=Pilkis S, el-Maghrabi M, Claus T |title=Fructose-2,6-bisphosphate in control of hepatic gluconeogenesis. From metabolites to molecular genetics |journal=Diabetes Care |volume=13 |issue=6 |pages=582-99 |year=1990 |pmid=2162755}}</ref> |
|||
Although fat is a common way of storing energy, in [[vertebrate]]s such as [[human]]s the [[fatty acid]]s in these stores cannot be converted to glucose through [[gluconeogenesis]] as these organisms cannot convert acetyl-CoA into [[pyruvate]].<ref name=Ensign>{{cite journal |author=Ensign S |title=Revisiting the glyoxylate cycle: alternate pathways for microbial acetate assimilation |journal=Mol Microbiol |volume=61 |issue=2 |pages=274-6 |year=2006 |pmid=16856935}}</ref> As a result, after long-term starvation, vertebrates need to produce [[ketone bodies]] from fatty acids to replace glucose in tissues such as the brain that cannot metabolize fatty acids.<ref>{{cite journal |author=Finn P, Dice J |title=Proteolytic and lipolytic responses to starvation |journal=Nutrition |volume=22 |issue=7–8 |pages=830-44 |year= |pmid=16815497}}</ref> In other organisms such as plants and bacteria, this metabolic problem is solved using the [[glyoxylate cycle]], which bypasses the [[decarboxylation]] step in the citric acid cycle and allows the transformation of acetyl-CoA to [[oxaloacetate]], where it can be used for the production of glucose.<ref name=Kornberg>{{cite journal |author=Kornberg H, Krebs H |title=Synthesis of cell constituents from C2-units by a modified tricarboxylic acid cycle |journal=Nature |volume=179 |issue=4568 |pages=988-91 |year=1957 |pmid=13430766}}</ref><ref name=Ensign/> |
|||
Polysaccharides and [[glycans]] are made by the sequential addition of monosaccharides by [[glycosyltransferase]] from a reactive sugar-phosphate donor such as [[uridine diphosphate glucose]] (UDP-glucose) to an acceptor [[hydroxyl]] group on the growing polysaccharide. As any of the [[hydroxyl]] groups on the ring of the substrate can be acceptors, the polysaccharides produced can have straight or branched structures.<ref>{{cite journal |author=Rademacher T, Parekh R, Dwek R |title=Glycobiology |journal=Annu Rev Biochem |volume=57 |issue= |pages=785–838 |year=1988 |pmid=3052290}}</ref> The polysaccharides produced can have structural or metabolic functions themselves, or be transferred to lipids and proteins by enzymes called [[oligosaccharyltransferase]]s.<ref>{{cite journal |author=Opdenakker G, Rudd P, Ponting C, Dwek R |title=Concepts and principles of glycobiology |url=http://www.fasebj.org/cgi/reprint/7/14/1330 |journal=FASEB J |volume=7 |issue=14 |pages=1330–7 |year=1993 |pmid=8224606}}</ref><ref>{{cite journal |author=McConville M, Menon A |title=Recent developments in the cell biology and biochemistry of glycosylphosphatidylinositol lipids (review) |journal=Mol Membr Biol |volume=17 |issue=1 |pages=1–16 |year=2000 |pmid=10824734}}</ref> |
|||
===Fatty acids, isoprenoids and steroids=== |
|||
{{further|[[Fatty acid synthesis]], [[mevalonate pathway]] and [[non-mevalonate pathway]]}} |
|||
[[Image:Sterol synthesis.svg|thumb|right|350px|Simplified version of the steroid synthesis pathway with the intermediates [[isopentenyl pyrophosphate]] (IPP), [[dimethylallyl pyrophosphate]] (DMAPP), [[geranyl pyrophosphate]] (GPP) and [[squalene]] shown. Some intermediates are omitted for clarity.]] |
|||
Fatty acids are made by [[fatty acid synthase]]s that polymerize and reduce acetyl-CoA units. The acyl chains in the fatty acids are extended by a cycle of reactions that add the actyl group, reduce it to the alcohol, [[dehydration reaction|dehydrate]] it to an [[alkene]] group and then reduce it again to an [[alkane]] group. The enzymes of fatty acid biosynthesis are divided into two groups, in animals and fungi all these fatty acid synthase reactions are carried out by a single multifunctional type I protein,<ref>{{cite journal |author=Chirala S, Wakil S |title=Structure and function of animal fatty acid synthase |journal=Lipids |volume=39 |issue=11 |pages=1045–53 |year=2004 |pmid=15726818}}</ref> while in plant [[plastid]]s and bacteria separate type II enzymes perform each step in the pathway.<ref>{{cite journal |author=White S, Zheng J, Zhang Y |title=The structural biology of type II fatty acid biosynthesis |journal=Annu Rev Biochem |volume=74 |issue= |pages=791–831 |year=2005 |pmid=15952903}}</ref><ref>{{cite journal |author=Ohlrogge J, Jaworski J |title=Regulation of fatty acid synthesis |journal=Annu Rev Plant Physiol Plant Mol Biol |volume=48 |issue= |pages=109–136 |year=1997 |pmid=15012259}}</ref> |
|||
[[Terpene]]s and [[terpenoid|isoprenoids]] are a large class of lipids that include the [[carotenoid]]s and form the largest class of plant [[natural product]]s.<ref>{{cite journal |author=Dubey V, Bhalla R, Luthra R |title=An overview of the non-mevalonate pathway for terpenoid biosynthesis in plants |url=http://www.ias.ac.in/jbiosci/sep2003/637.pdf |journal=J Biosci |volume=28 |issue=5 |pages=637-46 |year=2003 |pmid=14517367}}</ref> These compounds are made by the assembly and modification of [[isoprene]] units donated from the reactive precursors [[isopentenyl pyrophosphate]] and [[dimethylallyl pyrophosphate]].<ref name=Kuzuyama>{{cite journal |author=Kuzuyama T, Seto H |title=Diversity of the biosynthesis of the isoprene units |journal=Nat Prod Rep |volume=20 |issue=2 |pages=171-83 |year=2003 |pmid=12735695}}</ref> These precursors can be made in different ways. In animals and archaea, the [[mevalonate pathway]] produces these compounds from acetyl-CoA,<ref>{{cite journal |author=Grochowski L, Xu H, White R |title=Methanocaldococcus jannaschii uses a modified mevalonate pathway for biosynthesis of isopentenyl diphosphate |url=http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=16621811 |journal=J Bacteriol |volume=188 |issue=9 |pages=3192–8 |year=2006 |pmid=16621811}}</ref> while in plants and bacteria the [[non-mevalonate pathway]] uses pyruvate and [[glyceraldehyde 3-phosphate]] as substrates.<ref>{{cite journal |author=Lichtenthaler H |title=The 1-Ddeoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants |journal=Annu Rev Plant Physiol Plant Mol Biol |volume=50 |issue= |pages=47–65 |year=1999 |pmid=15012203}}</ref><ref name=Kuzuyama/> One important reaction that uses these activated isoprene donors is [[steroid]] biosynthesis. Here, the isoprene units are joined together to make [[squalene]] and then folded up and formed into a set of rings to make [[lanosterol]].<ref name=Schroepfer>{{cite journal |author=Schroepfer G |title=Sterol biosynthesis |journal=Annu Rev Biochem |volume=50 |issue= |pages=585–621 |year=1981 |pmid=7023367}}</ref> Lanosterol can then be converted into other steroids such as [[cholesterol]] and [[ergosterol]].<ref>{{cite journal |author=Lees N, Skaggs B, Kirsch D, Bard M |title=Cloning of the late genes in the ergosterol biosynthetic pathway of Saccharomyces cerevisiae—a review |journal=Lipids |volume=30 |issue=3 |pages=221-6 |year=1995 |pmid=7791529}}</ref><ref name=Schroepfer/> |
|||
===Proteins=== |
|||
{{further|[[Protein biosynthesis]], [[Amino acid synthesis]]}} |
|||
Organisms vary in their ability to synthesize the 20 common amino acids. Most bacteria and plants can synthesize all twenty, but mammals can synthesize only the ten nonessential amino acids.<ref name=Nelson/> Thus, the [[essential amino acid]]s must be obtained from food. All amino acids are synthesized from intermediates in glycolysis, the citric acid cycle, or the pentose phosphate pathway. Nitrogen is provided by [[glutamate]] and [[glutamine]]. Amino acid synthesis depends on the formation of the appropriate alpha-keto acid, which is then [[Transaminase|transaminated]] to form an amino acid.<ref>{{cite book | last = Guyton | first = Arthur C. | coauthors = John E. Hall | title = Textbook of Medical Physiology | publisher = Elsevier | date = 2006 | location = Philadelphia | pages = 855-6 | isbn = 0-7216-0240-1}}</ref> |
|||
Amino acids are made into proteins by being joined together in a chain by [[peptide bond]]s. Each different protein has a unique sequence of amino acid residues: this is its [[primary structure]]. Just as the letters of the alphabet can be combined to form an almost endless variety of words, amino acids can be linked in varying sequences to form a huge variety of proteins. Proteins are made from amino acids that have been activated by attachment to a [[transfer RNA]] molecule through an [[ester]] bond. This aminoacyl-tRNA precursor is produced in an [[Adenosine triphosphate|ATP]]-dependent reaction carried out by an [[aminoacyl tRNA synthetase]].<ref>{{cite journal | author = Ibba M, Söll D | title = The renaissance of aminoacyl-tRNA synthesis | url=http://www.molcells.org/home/journal/include/downloadPdf.asp?articleuid={A158E3B4-2423-4806-9A30-4B93CDA76DA0} | journal = EMBO Rep | volume = 2 | issue = 5 | pages = 382-7 | year = 2001 | pmid = 11375928}}</ref> This aminoacyl-tRNA is then a substrate for the [[ribosome]], which joins the amino acid onto the elongating protein chain, using the sequence information in a [[messenger RNA]].<ref>{{cite journal | author = Lengyel P, Söll D | title = Mechanism of protein biosynthesis | url=http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=378322&blobtype=pdf | journal = Bacteriol Rev | volume = 33 | issue = 2 | pages = 264–301 | year = 1969 | pmid = 4896351}}</ref> |
|||
===Nucleotide synthesis and salvage=== |
|||
{{further|[[Nucleotide salvage]], [[Pyrimidine biosynthesis]], and [[Purine#Metabolism|Purine metabolism]]}} |
|||
Nucleotides are made from amino acids, carbon dioxide and [[formic acid]] in pathways that require large amounts of metabolic energy.<ref name=Rudolph>{{cite journal |author=Rudolph F |title=The biochemistry and physiology of nucleotides |journal=J Nutr |volume=124 |issue=1 Suppl |pages=124S-127S |year=1994 |pmid=8283301}} {{cite journal |author=Zrenner R, Stitt M, Sonnewald U, Boldt R |title=Pyrimidine and purine biosynthesis and degradation in plants |journal=Annu Rev Plant Biol |volume=57 |issue= |pages=805-36 |year=2006 |pmid=16669783}}</ref> Consequently, most organisms have efficient systems to salvage preformed nucleotides.<ref name=Rudolph/><ref>{{cite journal |author=Stasolla C, Katahira R, Thorpe T, Ashihara H |title=Purine and pyrimidine nucleotide metabolism in higher plants |journal=J Plant Physiol |volume=160 |issue=11 |pages=1271–95 |year=2003 |pmid=14658380}}</ref> [[Purine]]s are synthesized as [[nucleoside]]s (bases attached to [[ribose]]). Both [[adenine]] and [[guanine]] are made from the precursor nucleoside [[inosine]] monophosphate, which is synthesized using atoms from the amino acids [[glycine]], [[glutamine]], and [[aspartic acid]], as well as [[formate]] transferred from the [[coenzyme]] [[folic acid|tetrahydrofolate]]. [[Pyrimidine]]s, on the other hand, are synthesized from the base [[Pyrimidinecarboxylic acid|orotate]], which is formed from glutamine and aspartate.<ref>{{cite journal |author=Smith J |title=Enzymes of nucleotide synthesis |journal=Curr Opin Struct Biol |volume=5 |issue=6 |pages=752-7 |year=1995 |pmid=8749362}}</ref> |
|||
==Xenobiotics and redox metabolism== |
|||
{{further|[[Detoxification]], [[Drug metabolism]] and [[Antioxidant]]s}} |
|||
All organisms are constantly exposed to compounds that they cannot use as foods and would be harmful if they accumulated in cells, as they have no metabolic function. These potentially damaging compounds are called [[xenobiotic]]s.<ref>{{cite journal |author=Testa B, Krämer S |title=The biochemistry of drug metabolism—an introduction: part 1. Principles and overview |journal=Chem Biodivers |volume=3 |issue=10 |pages=1053-101 |year=2006 |pmid=17193224}}</ref> Xenobiotics such as [[drug|synthetic drugs]], [[poison|natural poisons]] and [[antibiotic]]s are detoxified by a set of xenobiotic-metabolizing enzymes. In humans, these include [[cytochrome P450|cytochrome P450 oxidase]]s,<ref>{{cite journal |author=Danielson P |title=The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans |journal=Curr Drug Metab |volume=3 |issue=6 |pages=561-97 |year=2002 |pmid=12369887}}</ref> [[Glucuronosyltransferase|UDP-glucuronosyltransferases]]s,<ref>{{cite journal |author=King C, Rios G, Green M, Tephly T |title=UDP-glucuronosyltransferases |journal=Curr Drug Metab |volume=1 |issue=2 |pages=143-61 |year=2000 |pmid=11465080}}</ref> and [[glutathione S-transferase|glutathione ''S''-transferase]]s.<ref>{{cite journal |author=Sheehan D, Meade G, Foley V, Dowd C |title=Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily |url=http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=11695986 |journal=Biochem J |volume=360 |issue=Pt 1 |pages=1–16 |year=2001 |pmid=11695986}}</ref> This system of enzymes acts in three stages to firstly oxidize the xenobiotic (phase I) and then conjugate water-soluble groups onto the molecule (phase II). The modified water-soluble xenobiotic can then be pumped out of cells and in multicellular organisms may be further metabolized before being excreted (phase III). In [[ecology]], these reactions are particularly important in microbial [[biodegradation]] of pollutants and the [[bioremediation]] of contaminated land and oil spills.<ref>{{cite journal |author=Galvão T, Mohn W, de Lorenzo V |title=Exploring the microbial biodegradation and biotransformation gene pool |journal=Trends Biotechnol |volume=23 |issue=10 |pages=497–506 |year=2005 |pmid=16125262}}</ref> Many of these microbial reactions are shared with multicellular organisms, but due to their incredible diversity, microbes are able to deal with a far wider range of xenobiotics than multicellular organisms and can degrade even [[persistent organic pollutant]]s such as [[organochloride]] compounds.<ref>{{cite journal |author=Janssen D, Dinkla I, Poelarends G, Terpstra P |title=Bacterial degradation of xenobiotic compounds: evolution and distribution of novel enzyme activities |journal=Environ Microbiol |volume=7 |issue=12 |pages=1868–82 |year=2005 |pmid=16309386}}</ref> |
|||
A related problem for [[aerobic organism]]s is [[oxidative stress]].<ref name=Davies>{{cite journal |author=Davies K |title=Oxidative stress: the paradox of aerobic life |journal=Biochem Soc Symp |volume=61 |issue= |pages=1–31 |year=1995 |pmid=8660387}}</ref> Here, processes including [[oxidative phosphorylation]] and the formation of [[disulfide bond]]s during [[protein folding]] produce [[reactive oxygen species]] such as [[hydrogen peroxide]].<ref>{{cite journal |author=Tu B, Weissman J |title=Oxidative protein folding in eukaryotes: mechanisms and consequences |url=http://www.jcb.org/cgi/content/full/164/3/341 |journal=J Cell Biol |volume=164 |issue=3 |pages=341-6 |year=2004 |pmid=14757749}}</ref> These damaging oxidants are removed by [[antioxidant]] metabolites such as [[glutathione]] and enzymes such as [[catalase]]s and [[peroxidase]]s.<ref name=Sies>{{cite journal |author=Sies H |title=Oxidative stress: oxidants and antioxidants |url=http://ep.physoc.org/cgi/reprint/82/2/291.pdf |journal=Exp Physiol |volume=82 |issue=2 |pages=291-5 |year=1997 |pmid=9129943}}</ref><ref name=Vertuani>{{cite journal |author=Vertuani S, Angusti A, Manfredini S |title=The antioxidants and pro-antioxidants network: an overview |journal=Curr Pharm Des |volume=10 |issue=14 |pages=1677–94 |year=2004 |pmid=15134565}}</ref> |
|||
==Thermodynamics of living organisms== |
|||
{{further|[[Biological thermodynamics]]}} |
|||
Living organisms must obey the [[laws of thermodynamics]]. The [[second law of thermodynamics]] states that in any [[closed system]], the amount of [[entropy]] (disorder) will tend to increase. Although living organisms' amazing complexity appears to contradict this law, life is possible as all organisms are [[open system (system theory)|open system]]s that exchange matter and energy with their surroundings. Thus living systems are not in [[Thermodynamic equilibrium|equilibrium]], but instead are [[dissipative system]]s that maintain their state of high complexity by causing a larger increase in the entropy of their environments.<ref>{{cite journal |author=von Stockar U, Liu J |title=Does microbial life always feed on negative entropy? Thermodynamic analysis of microbial growth |journal=Biochim Biophys Acta |volume=1412 |issue=3 |pages=191–211 |year=1999 |pmid=10482783}}</ref> The metabolism of a cell achieves this by coupling the [[spontaneous process]]es of catabolism to the non-spontaneous processes of anabolism. In [[non-equilibrium thermodynamics|thermodynamic]] terms, metabolism maintains order by creating disorder.<ref>{{cite journal |author=Demirel Y, Sandler S |title=Thermodynamics and bioenergetics |journal=Biophys Chem |volume=97 |issue=2–3 |pages=87–111 |year=2002 |pmid=12050002}}</ref> |
|||
==Regulation and control== |
|||
{{further|[[Metabolic pathway]], [[metabolic control analysis]], [[hormone]] and [[cell signaling]]}} |
|||
As the environments of most organisms are constantly changing, the reactions of metabolism must be finely [[Control theory|regulated]] to maintain a constant set of conditions within cells, a condition called [[homeostasis]].<ref>{{cite journal |author=Albert R |title=Scale-free networks in cell biology |url=http://jcs.biologists.org/cgi/content/full/118/21/4947 |journal=J Cell Sci |volume=118 |issue=Pt 21 |pages=4947–57 |year=2005 |pmid=16254242}}</ref><ref>{{cite journal |author=Brand M |title=Regulation analysis of energy metabolism |url=http://jeb.biologists.org/cgi/reprint/200/2/193 |journal=J Exp Biol |volume=200 |issue=Pt 2 |pages=193–202 |year=1997 |pmid=9050227}}</ref> Metabolic regulation also allows organisms to respond to signals and interact actively with their environments.<ref>{{cite journal |author=Soyer O, Salathé M, Bonhoeffer S |title=Signal transduction networks: topology, response and biochemical processes |journal=J Theor Biol |volume=238 |issue=2 |pages=416-25 |year=2006 |pmid=16045939}}</ref> Two closely-linked concepts are important for understanding how metabolic pathways are controlled. Firstly, the ''regulation'' of an enzyme in a pathway is how its activity is increased and decreased in response to signals. Secondly, the ''control'' exerted by this enzyme is the effect that these changes in its activity have on the overall rate of the pathway (the [[flux]] through the pathway).<ref name=Salter>{{cite journal |author=Salter M, Knowles R, Pogson C |title=Metabolic control |journal=Essays Biochem |volume=28 |issue= |pages=1–12 |year=1994 |pmid=7925313}}</ref> For example, an enzyme may show large changes in activity (''i.e.'' it is highly regulated) but if these changes have little effect on the flux of a metabolic pathway, then this enzyme is not involved in the control of the pathway.<ref>{{cite journal |author=Westerhoff H, Groen A, Wanders R |title=Modern theories of metabolic control and their applications (review) |journal=Biosci Rep |volume=4 |issue=1 |pages=1–22 |year=1984 |pmid=6365197}}</ref> |
|||
[[Image:Insulin_glucose_metabolism_ZP.svg|right|thumbnail|400px|'''Effect of insulin on glucose uptake and metabolism.''' Insulin binds to its receptor (1) which in turn starts many protein activation cascades (2). These include: translocation of Glut-4 transporter to the [[plasma membrane]] and influx of glucose (3), [[glycogen]] synthesis (4), [[glycolysis]] (5) and [[fatty acid]] synthesis (6).]] |
|||
There are multiple levels of metabolic regulation. In intrinsic regulation, the metabolic pathway self-regulates to respond to changes in the levels of substrates or products; for example, a decrease in the amount of product can increase the [[flux]] through the pathway to compensate.<ref name=Salter/> This type of regulation often involves [[allosteric regulation]] of the activities of multiple enzymes in the pathway.<ref>{{cite journal |author=Fell D, Thomas S |title=Physiological control of metabolic flux: the requirement for multisite modulation |url=http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=7575476 |journal=Biochem J |volume=311 (Pt 1) |issue= |pages=35-9 |year=1995 |pmid=7575476}}</ref> Extrinsic control involves a cell in a multicellular organism changing its metabolism in response to signals from other cells. These signals are usually in the form of soluble messengers such as [[hormone]]s and [[growth factor]]s and are detected by specific [[receptor (biochemistry)|receptors]] on the cell surface.<ref>{{cite journal |author=Hendrickson W |title=Transduction of biochemical signals across cell membranes |journal=Q Rev Biophys |volume=38 |issue=4 |pages=321-30 |year=2005 |pmid=16600054}}</ref> These signals are then transmitted inside the cell by [[second messenger system]]s that often involved the [[phosphorylation]] of proteins.<ref>{{cite journal |author=Cohen P |title=The regulation of protein function by multisite phosphorylation—a 25 year update |journal=Trends Biochem Sci |volume=25 |issue=12 |pages=596–601 |year=2000 |pmid=11116185}}</ref> |
|||
A very well understood example of extrinsic control is the regulation of glucose metabolism by the hormone [[insulin]].<ref>{{cite journal |author=Lienhard G, Slot J, James D, Mueckler M |title=How cells absorb glucose |journal=Sci Am |volume=266 |issue=1 |pages=86–91 |year=1992 |pmid=1734513}}</ref> Insulin is produced in response to rises in [[blood sugar|blood glucose levels]]. Binding of the hormone to [[insulin receptor]]s on cells then activates a cascade of [[protein kinase]]s that cause the cells to take up glucose and convert it it into storage molecules such as fatty acids and [[glycogen]].<ref>{{cite journal |author=Roach P |title=Glycogen and its metabolism |journal=Curr Mol Med |volume=2 |issue=2 |pages=101-20 |year=2002 |pmid=11949930}}</ref> The metabolism of glycogen is controlled by activity of [[phosphorylase]], the enzyme that breaks down glycogen, and [[glycogen synthase]], the enzyme that makes it. These enzymes are regulated in a reciprocal fashion, with phosphorylation inhibiting glycogen synthase, but activating phosphorylase. Insulin causes glycogen synthesis by activating [[phosphatase|protein phosphatase]]s and producing a decrease in the phosphorylation of these enzymes.<ref>{{cite journal |author=Newgard C, Brady M, O'Doherty R, Saltiel A |title=Organizing glucose disposal: emerging roles of the glycogen targeting subunits of protein phosphatase-1 |url=http://diabetes.diabetesjournals.org/cgi/reprint/49/12/1967.pdf |journal=Diabetes |volume=49 |issue=12 |pages=1967–77 |year=2000 |pmid=11117996}}</ref> |
|||
==Evolution== |
|||
{{further|[[Molecular evolution]] and [[Phylogenetics]]}} |
|||
[[Image:Tree of life 1500px coloured.png|thumb|left|400px|[[Phylogenetic tree|Evolutionary tree]] showing the common ancestry of organisms from all three [[Domain (biology)|domains]] of life. [[Bacteria]] are colored blue, [[eukaryote]]s red, and [[archaea]] green. Relative positions of some of the [[phylum|phyla]] included are shown around the tree.]] |
|||
The central pathways of metabolism described above, such as glycolysis and the citric acid cycle, are present in all [[Three-domain system|three domains]] of living things and were present in the [[last universal ancestor]].<ref>{{cite journal |author=Romano A, Conway T |title=Evolution of carbohydrate metabolic pathways |journal=Res Microbiol |volume=147 |issue=6–7 |pages=448-55 |year= |pmid=9084754}}</ref><ref name=SmithE/> This universal ancestral cell was [[prokaryote|prokaryotic]] and probably a [[methanogen]] that had extensive amino acid, nucleotide, carbohydrate and lipid metabolism.<ref>{{cite journal |author=Koch A |title=How did bacteria come to be? |journal=Adv Microb Physiol |volume=40 |issue= |pages=353-99 |year=1998 |pmid=9889982}} {{cite journal |author=Ouzounis C, Kyrpides N |title=The emergence of major cellular processes in evolution |journal=FEBS Lett |volume=390 |issue=2 |pages=119-23 |year=1996 |pmid=8706840}}</ref> The retention of these ancient pathways during later [[evolution]] may be the result of these reactions being an optimal solution to their particular metabolic problems, with pathways such as glycolysis and the citric acid cycle producing their end products highly efficiently and in a minimal number of steps.<ref name=Ebenhoh/><ref name=Cascante/> |
|||
Many models have been proposed to describe the mechanisms by which novel metabolic pathways evolve. These include the sequential addition of novel enzymes to a short ancestral pathway, the duplication and then divergence of entire pathways as well as the recruitment of pre-existing enzymes and their assembly into a novel reaction pathway.<ref>{{cite journal |author=Schmidt S, Sunyaev S, Bork P, Dandekar T |title=Metabolites: a helping hand for pathway evolution? |journal=Trends Biochem Sci |volume=28 |issue=6 |pages=336-41 |year=2003 |pmid=12826406}}</ref> The relative importance of these mechanisms is unclear, but genomic studies have shown that enzymes in a pathway are likely to have a shared ancestry, suggesting that many pathways have evolved in a step-by-step fashion with novel functions being created from pre-existing steps in the pathway.<ref>{{cite journal |author=Light S, Kraulis P |title=Network analysis of metabolic enzyme evolution in Escherichia coli |journal=BMC Bioinformatics |volume=5 |issue= |pages=15 |year= |pmid=15113413}} {{cite journal |author=Alves R, Chaleil R, Sternberg M |title=Evolution of enzymes in metabolism: a network perspective |journal=J Mol Biol |volume=320 |issue=4 |pages=751-70 |year=2002 |pmid=12095253}}</ref> Another possibility is that some parts of metabolism might exist as "modules" that can be reused in different pathways and perform similar functions on different molecules.<ref>{{cite journal |author=Spirin V, Gelfand M, Mironov A, Mirny L |title=A metabolic network in the evolutionary context: multiscale structure and modularity |url=http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=16731630 |journal=Proc Natl Acad Sci U S A |volume=103 |issue=23 |pages=8774–9 |year=2006 |pmid=16731630}}</ref> |
|||
The evolution of organisms can also produce the loss of metabolic pathways. For example, in some [[parasite]]s metabolic processes that are not essential for survival are lost and preformed amino acids, nucleotides and carbohydrates may instead be scavenged from the [[host (biology)|host]].<ref>{{cite journal |author=Lawrence J |title=Common themes in the genome strategies of pathogens |journal=Curr Opin Genet Dev |volume=15 |issue=6 |pages=584-8 |year=2005 |pmid=16188434}} {{cite journal |author=Wernegreen J |title=For better or worse: genomic consequences of intracellular mutualism and parasitism |journal=Curr Opin Genet Dev |volume=15 |issue=6 |pages=572-83 |year=2005 |pmid=16230003}}</ref> Similar reduced metabolic capabilities are seen in [[endosymbiont|endosymbiotic]] organisms.<ref>{{cite journal |author=Pál C, Papp B, Lercher M, Csermely P, Oliver S, Hurst L |title=Chance and necessity in the evolution of minimal metabolic networks |journal=Nature |volume=440 |issue=7084 |pages=667-70 |year=2006 |pmid=16572170}}</ref> |
|||
==Investigation and manipulation== |
|||
{{further|[[Protein methods]], [[proteomics]], [[metabolomics]] and [[metabolic network modelling]]}} |
|||
[[Image:A thaliana metabolic network.png|thumb|300px|right|[[Metabolic network]] of the ''[[Arabidopsis thaliana]]'' [[citric acid cycle]]. [[Enzyme]]s and [[metabolomics|metabolite]]s are shown as red squares and the interactions between them as black lines.]] |
|||
Classically, metabolism is studied by a [[reductionism|reductionist]] approach that focuses on a single metabolic pathway. Particularly valuable is the use of [[radioactive tracer]]s at the whole-organism, tissue and cellular levels, which define the paths from precursors to final products by identifying radioactively-labelled intermediates and products.<ref>{{cite journal |author=Rennie M |title=An introduction to the use of tracers in nutrition and metabolism |journal=Proc Nutr Soc |volume=58 |issue=4 |pages=935-44 |year=1999 |pmid=10817161}}</ref> The enzymes that catalyze these chemical reactions can then be [[protein purification|purified]] and their [[enzyme kinetics|kinetics]] and responses to [[enzyme inhibitor|inhibitors]] investigated. A parallel approach is to identify the small molecules in a cell or tissue; the complete set of these molecules is called the [[metabolome]]. Overall, these studies give a good view of the structure and function of simple metabolic pathways, but are inadequate when applied to more complex systems such as the metabolism of a complete cell.<ref>{{cite journal |author=Phair R |title=Development of kinetic models in the nonlinear world of molecular cell biology |journal=Metabolism |volume=46 |issue=12 |pages=1489–95 |year=1997 |pmid=9439549}}</ref> |
|||
An idea of the complexity of the [[metabolic network]]s in cells that contain thousands of different enzymes is given by the figure showing the interactions between just 43 proteins and 40 metabolites to the right: the sequences of genomes provide lists containing anything up to 45,000 genes.<ref>{{cite journal |author=Sterck L, Rombauts S, Vandepoele K, Rouzé P, Van de Peer Y |title=How many genes are there in plants (... and why are they there)? |journal=Curr Opin Plant Biol |volume=10 |issue=2 |pages=199–203 |year=2007 |pmid=17289424}}</ref> However, it is now possible to use this genomic data to reconstruct complete networks of biochemical reactions and produce more [[Holism|holistic]] mathematical models that may explain and predict their behavior.<ref>{{cite journal |author=Borodina I, Nielsen J |title=From genomes to in silico cells via metabolic networks |journal=Curr Opin Biotechnol |volume=16 |issue=3 |pages=350-5 |year=2005 |pmid=15961036}}</ref> These models are especially powerful when used to integrate the pathway and metabolite data obtained through classical methods with data on [[gene expression]] from [[proteomics|proteomic]] and [[DNA microarray]] studies.<ref>{{cite journal |author=Gianchandani E, Brautigan D, Papin J |title=Systems analyses characterize integrated functions of biochemical networks |journal=Trends Biochem Sci |volume=31 |issue=5 |pages=284-91 |year=2006 |pmid=16616498}}</ref> |
|||
A major technological application of this information is [[metabolic engineering]]. Here, organisms such as [[yeast]], [[plant]]s or [[bacteria]] are genetically-modified to make them more useful in [[biotechnology]] and aid the production of [[drug]]s such as [[antibiotic]]s or industrial chemicals such as [[1,3-Propanediol|1,3-propanediol]] and [[shikimic acid]].<ref>{{cite journal |author=Thykaer J, Nielsen J |title=Metabolic engineering of beta-lactam production |journal=Metab Eng |volume=5 |issue=1 |pages=56–69 |year=2003 |pmid=12749845}} |
|||
{{cite journal |author=González-Pajuelo M, Meynial-Salles I, Mendes F, Andrade J, Vasconcelos I, Soucaille P |title=Metabolic engineering of Clostridium acetobutylicum for the industrial production of 1,3-propanediol from glycerol |journal=Metab Eng |volume=7 |issue=5–6 |pages=329-36 |year= |pmid=16095939}} |
|||
{{cite journal |author=Krämer M, Bongaerts J, Bovenberg R, Kremer S, Müller U, Orf S, Wubbolts M, Raeven L |title=Metabolic engineering for microbial production of shikimic acid |journal=Metab Eng |volume=5 |issue=4 |pages=277-83 |year=2003 |pmid=14642355}}</ref> These genetic modifications usually aim to reduce the amount of energy used to produce the product, increase yields and reduce the production of wastes.<ref>{{cite journal |author=Koffas M, Roberge C, Lee K, Stephanopoulos G |title=Metabolic engineering |journal=Annu Rev Biomed Eng |volume=1 |issue= |pages=535-57 |year=1999 |pmid=11701499}}</ref> |
|||
== History == |
|||
{{further|[[History of biochemistry]] and [[History of molecular biology]]}} |
|||
[[Image:SantoriosMeal.jpg|thumb|right|150px|[[Santorio Santorio]] in his steelyard balance, from ''Ars de statica medecina'', first published 1614]] |
|||
The term ''metabolism'' is derived from the [[Greek language|Greek]] Μεταβολισμός – "Metabolismos" for "change", or "overthrow".<ref>{{cite web | title=Metabolism |publisher=The Online Etymology Dictionary | url=http://www.etymonline.com/index.php?term=metabolism |accessdate=2007-02-20}}</ref> The history of the scientific study of metabolism spans 400 years and has moved from examining whole animals in early studies, to examining individual metabolic reactions in modern biochemistry. The first controlled experiments in human metabolism were published by [[Santorio Santorio]] in [[1614]] in his book ''Ars de statica medecina''.<ref>{{cite journal |author=Eknoyan G |title=Santorio Sanctorius (1561–1636) - founding father of metabolic balance studies |journal=Am J Nephrol |volume=19 |issue=2 |pages=226-33 |year=1999 |pmid=10213823}}</ref> He described how he weighed himself before and after eating, sleeping, working, sex, fasting, drinking, and excreting. He found that most of the food he took in was lost through what he called "insensible perspiration". |
|||
In these early studies, the mechanisms of these metabolic processes had not been identified and a [[vitalism|vital force]] was thought to animate living tissue.<ref>Williams, H. S. (1904) [http://etext.lib.virginia.edu/toc/modeng/public/Wil4Sci.html A History of Science: in Five Volumes. Volume IV: Modern Development of the Chemical and Biological Sciences] Harper and Brothers (New York) Retrieved on 2007-03-26</ref> In the 19th century, when studying the [[fermentation (food)|fermentation]] of sugar to [[alcohol]] by [[yeast]], [[Louis Pasteur]] concluded that fermentation was catalyzed by substances within the yeast cells he called "ferments". He wrote that "alcoholic fermentation is an act correlated with the life and organization of the yeast cells, not with the death or putrefaction of the cells."<ref>{{cite journal |author=Dubos J.|year= 1951|title= Louis Pasteur: Free Lance of Science, Gollancz. Quoted in Manchester K. L. (1995) Louis Pasteur (1822 – 1895)—chance and the prepared mind.|journal= Trends Biotechnol| volume=13 |issue=12 |pages=511–515 |id= PMID 8595136}}</ref> This discovery, along with the publication by [[Friedrich Woehler|Friedrich Wöhler]] in [[1828]] of the chemical synthesis of [[urea]],<ref>{{cite journal |author=Kinne-Saffran E, Kinne R |title=Vitalism and synthesis of urea. From Friedrich Wöhler to Hans A. Krebs |journal=Am J Nephrol |volume=19 |issue=2 |pages=290-4 |year=1999 |pmid=10213830}}</ref> proved that the organic compounds and chemical reactions found in cells were no different in principle than any other part of chemistry. |
|||
It was the discovery of [[enzyme]]s at the beginning of the 20th century by [[Eduard Buchner]] that separated the study of the chemical reactions of metabolism from the biological study of cells, and marked the beginnings of [[biochemistry]].<ref>Eduard Buchner's 1907 [http://nobelprize.org/nobel_prizes/chemistry/laureates/1907/buchner-lecture.html Nobel lecture] at http://nobelprize.org Accessed 2007-03-20</ref> The mass of biochemical knowledge grew rapidly throughout the early 20th century. One of the most prolific of these modern biochemists was [[Hans Adolf Krebs|Hans Krebs]] who made huge contributions to the study of metabolism.<ref>{{cite journal |author=Kornberg H |title=Krebs and his trinity of cycles |journal=Nat Rev Mol Cell Biol |volume=1 |issue=3 |pages=225-8 |year=2000 |pmid=11252898}}</ref> He discovered the urea cycle and later, working with [[Hans Kornberg]], the citric acid cycle and the glyoxylate cycle.<ref>Krebs H A, Henseleit K (1932) "Untersuchungen über die Harnstoffbildung im tierkorper." ''Z. Physiol. Chem.'' 210, 33 – 66. {{cite journal |author=Krebs H, Johnson W |title=Metabolism of ketonic acids in animal tissues |url=http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=16746382 |journal=Biochem J |volume=31 |issue=4 |pages=645-60 |year=1937 |pmid=16746382}}</ref><ref name=Kornberg/> Modern biochemical research has been greatly aided by the development of new techniques such as [[chromatography]], [[X-ray diffraction]], [[protein nuclear magnetic resonance spectroscopy|NMR spectroscopy]], [[radioisotopic labelling]], [[electron microscope|electron microscopy]] and [[molecular dynamics]] simulations. These techniques have allowed the discovery and detailed analysis of the many molecules and metabolic pathways in cells. |
|||
== See also == |
|||
{{wikibooks}} |
|||
{{Wikiversity|Topic:Biochemistry}} |
|||
{{portalpar|Metabolism}} |
|||
* [[Inborn error of metabolism]] |
|||
* [[Basal metabolic rate]] |
|||
* [[Thermic effect of food]] |
|||
* [[Iron-sulfur world theory]], a "metabolism first" theory of the [[origin of life]]. |
|||
* [[Calorimetry]] |
|||
* [[Respirometry]] |
|||
* [[Anthropogenic metabolism]] |
|||
== References == |
|||
<div class="reflist4" style="height: 220px; overflow: auto; padding: 3px" > |
|||
<references/> |
|||
</div> |
|||
==Further reading== |
|||
'''Introductory''' |
|||
* Rose, S. and Mileusnic, R., ''The Chemistry of Life.'' (Penguin Press Science, 1999), ISBN 0-14027-273-9 |
|||
* Schneider, E. D. and Sagan, D., ''Into the Cool: Energy Flow, Thermodynamics, and Life.'' (University Of Chicago Press, 2005), ISBN 0-22673-936-8 |
|||
* Lane, N., ''Oxygen: The Molecule that Made the World.'' (Oxford University Press, USA, 2004), ISBN 0-19860-783-0 |
|||
'''Advanced''' |
|||
* Price, N. and Stevens, L., ''Fundamentals of Enzymology: Cell and Molecular Biology of Catalytic Proteins.'' (Oxford University Press, 1999), ISBN 0-19850-229-X |
|||
* Berg, J. Tymoczko, J. and Stryer, L., ''Biochemistry.'' (W. H. Freeman and Company, 2002), ISBN 0-71674-955-6 |
|||
* Cox, M. and Nelson, D. L., ''Lehninger Principles of Biochemistry.'' (Palgrave Macmillan, 2004), ISBN 0-71674-339-6 |
|||
* Brock, T. D. Madigan, M. T. Martinko, J. and Parker J., ''Brock's Biology of Microorganisms.'' (Benjamin Cummings, 2002), ISBN 0-13066-271-2 |
|||
* Da Silva, J.J.R.F. and Williams, R. J. P., ''The Biological Chemistry of the Elements: The Inorganic Chemistry of Life.'' (Clarendon Press, 1991), ISBN 0-19855-598-9 |
|||
* Nicholls, D. G. and Ferguson, S. J., ''Bioenergetics.'' (Academic Press Inc., 2002), ISBN 0-12518-121-3 |
|||
==<font color=#FFFFFF>External links</font>== |
|||
<div style="clear: both; width: 100%; padding: 0; text-align: left; border: none;" class="NavFrame"> |
|||
<div style="background: #ccddcc; text-align: center; border: 1px solid #667766" class="NavHead">'''External links''' |
|||
</div> |
|||
<div class="NavContent"> |
|||
{| class="toccolours" style="width: 100%; border-top: none;" |
|||
'''General information''' |
|||
* [http://www2.ufp.pt/~pedros/bq/integration.htm Interactive Flow Chart of the Major Metabolic Pathways] |
|||
* [http://www.biochemweb.org/metabolism.shtml Metabolism, Cellular Respiration and Photosynthesis] The Virtual Library of Biochemistry and Cell Biology at biochemweb.org |
|||
* [http://www.rpi.edu/dept/bcbp/molbiochem/MBWeb/mb1/MB1index.html The Biochemistry of Metabolism] |
|||
* [http://www.stthomas.edu/biol/ecophys/homepage/homepage.html Advanced Animal Metabolism Calculators/ Interactive Learning Tools] |
|||
* [http://www.slic2.wsu.edu:82/hurlbert/micro101/pages/Chap7.html Microbial metabolism] Simple overview. School level. |
|||
* [http://www.gwu.edu/~mpb/ Metabolic Pathways of Biochemistry] Graphical representations of major metabolic pathways. |
|||
* [http://www.chemsoc.org/networks/LearnNet/cfb/contents.htm Chemistry for biologists] Introduction to the chemistry of metabolism. School level. |
|||
* [http://www.sparknotes.com/testprep/books/sat2/biology/chapter5section7.rhtml Sparknotes SAT biochemistry] Overview of biochemistry. School level. |
|||
* [http://web.mit.edu/esgbio/www/7001main.html MIT Biology Hypertextbook] Undergraduate-level guide to molecular biology. |
|||
* [http://www.britannica.com/eb/article-9109831/metabolism Article on metabolism at The Encyclopœdia Britannica] Concentrates on human metabolism (Free access). |
|||
'''Glossaries and dictionaries''' |
|||
* [http://biology.clc.uc.edu/scripts/glossary.pl Glossary of biochemical terms] |
|||
* [http://www.portlandpress.com/pp/books/online/glick/default.htm Glossary of biochemical terms] |
|||
* [http://www.biology-online.org/dictionary.asp On-line biology dictionary] |
|||
'''Human metabolism''' |
|||
* [http://library.med.utah.edu/NetBiochem/titles.htm Topics in Medical Biochemistry] Guide to human metabolic pathways. School level. |
|||
* [http://www.indstate.edu/thcme/mwking/ THE Medical Biochemistry Page] Comprehensive resource on human metabolism. |
|||
'''Databases''' |
|||
* [http://www.expasy.org/cgi-bin/show_thumbnails.pl Flow Chart of Metabolic Pathways] at [[ExPASy]] |
|||
* [http://www.genome.ad.jp/kegg/ The KEGG PATHWAY Database] |
|||
* [http://www.sigmaaldrich.com/img/assets/4202/MetabolicPathways_6_17_04_.pdf IUBMB-Nicholson Metabolic Pathways Chart] |
|||
* [http://www.reactome.org/ Reactome - a knowledgebase of biological processes] |
|||
'''Metabolic pathways''' |
|||
* [http://biotech.icmb.utexas.edu/glycolysis/glycohome.html Guide to Glycolysis] School level. |
|||
* [http://helios.bto.ed.ac.uk/bto/microbes/nitrogen.htm The Nitrogen cycle and Nitrogen fixation] |
|||
* [http://www.oxygraphics.co.uk/cds.htm Downloadable guide to photosynthesis] School level. |
|||
* [http://photoscience.la.asu.edu/photosyn/education/learn.html What is Photosynthesis?] Collection of photosynthesis articles and resources. |
|||
|} |
|||
</div> |
|||
</div> |
|||
''</s></s> |
|||
--> |
|||
==Anabolismo e catabolismo== |
==Anabolismo e catabolismo== |
||
O metabolismo é normalmente dividido em dois grupo: '''[[anabolismo]]''' e '''[[catabolismo]]'''. Reacções anabólicas, ou reacções de síntese (produção de nova matéria), são reacções químicas que produzem nova matéria [[composto orgânico|orgânica]] nos seres vivos. Sintetizam-se novos [[composto químico|compostos]] ([[molécula]]s mais complexas) a partir de moléculas mais simples (com consumo de [[ATP]]). Reacções catabólicas, ou reacções de decomposição/degradação, são reacções químicas que produzem grandes quantidades de energia livre (sob a forma de [[ATP]]) a partir da decomposição ou degradação de moléculas mais complexas (matéria orgânica). |
O metabolismo é normalmente dividido em dois grupo: '''[[anabolismo]]''' e '''[[catabolismo]]'''. Reacções anabólicas, ou reacções de síntese (produção de nova matéria), são reacções químicas que produzem nova matéria [[composto orgânico|orgânica]] nos seres vivos. Sintetizam-se novos [[composto químico|compostos]] ([[molécula]]s mais complexas) a partir de moléculas mais simples (com consumo de [[ATP]]). Reacções catabólicas, ou reacções de decomposição/degradação, são reacções químicas que produzem grandes quantidades de energia livre (sob a forma de [[ATP]]) a partir da decomposição ou degradação de moléculas mais complexas (matéria orgânica). |
||
| Linha 11: | Linha 304: | ||
Quando o catabolismo supera em atividade o anabolismo, o organismo perde [[peso]], o que acontece em períodos de [[jejum]] ou [[doença]]; mas se o anabolismo superar o catabolismo, o organismo cresce ou ganha peso. Se ambos os processos estão em equilíbrio, o organismo encontra-se em [[equilíbrio dinâmico]] ou [[homeostase]]. |
Quando o catabolismo supera em atividade o anabolismo, o organismo perde [[peso]], o que acontece em períodos de [[jejum]] ou [[doença]]; mas se o anabolismo superar o catabolismo, o organismo cresce ou ganha peso. Se ambos os processos estão em equilíbrio, o organismo encontra-se em [[equilíbrio dinâmico]] ou [[homeostase]]. |
||
==Referências== |
|||
{{Reflist|2}} |
|||
=={{Links Externos}}== |
=={{Links Externos}}== |
||
{{portal-bioquímica}} |
{{portal-bioquímica}} |
||
Revisão das 18h32min de 24 de junho de 2007
| Parte ou a integralidade do conteúdo desta página resulta da tradução de uma página originalmente presente numa Wikipédia noutra língua. A página correspondente pode ser conferida aqui. As fontes não foram verificadas. |

Metabolismo (do grego metabolé, μεταβολισμος, que significa "mudança", troca, acrescido de ismo) é o conjunto de transformações que as substâncias químicas sofrem no interior dos organismos vivos. É também usado o termo "metabolismo celular" referindo-se mais especificamente ao conjunto de todas as reacções químicas que ocorrem nas células. Estas reações químicas são responsáveis pelos processos de síntese e degradação dos nutrientes na célula.
Estes processos são a base da vida, permitindo o crescimento e reprodução das células, mantendo as suas estruturas e adequando respostas aos seus ambientes.
As reacções químicas do metabolismo estão organizadas em vias metabólicas, que são sequências de reacções em que o produto de uma reacção é utilizado como reagente na reacção seguinte. Diferentes enzimas catalisam diferentes passos de vias metabólicas, agindo de forma concertada de modo a não interromper o fluxo nessas vias. As enzimas são vitais para o metabolismo porque permitem a realização de reacções desejáveis mas termodinamicamente desfavoráveis, ao acoplá-las a reacções mais favoráveis. As enzimas regulam as vias metabólicas em resposta a mudanças no ambiente celular ou a sinais de outras células.
O metabolismo de um organismo determina que substâncias são nutricionais e quais são tóxicas. Por exemplo, alguns procariontes utilizam ácido sulfídrico como nutriente; este gás é no entanto venenoso para animais[1]. A velocidade a que se processa o metabolismo, determinada pela taxa metabólica, também influencia a quantidade de alimento requerida por um organismo.
Uma característica do metabolismo é a semelhança de vias metabólicas básicas entre espécies muito diferentes. Por exemplo, o conjunto de intermediários reaccionais encontrados no ciclo dos ácidos tricarboxílicos é encontrado de forma universal, em células tão diferentes como a bactéria Escherichia coli ou o elefante[2]. Esta estrutura metabólica semelhante está provavelmente associada à grande eficiência dessas vias e na sua antiguidade na história da evolução[3][4].
O metabolismo é fundamentalmente estudado pela Bioquímica, usando muitas vezes também técnicas ligadas à Biologia Molecular e à Genética.
Substâncias bioquímicas relevantes
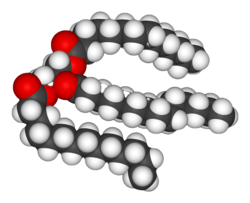
A maioria das estruturas que compõem os seres vivos é fabricada a partir de três classes básicas de moléculas: aminoácidos, glícidos e lípidos. Como estas moléculas são vitais, o metabolismo concentra-se no fabrico destas, na construção de células e tecidos ou na sua degradação para uso como fonte de energia. Muitos compostos bioquímicos podem ser condensados formando polímeros, como o ADN e as proteínas. Estas macromoléculas são parte essencial de todos os organismos vivos.
Alguns dos polímeros mais comuns estão listados abaixo:
| Tipo de molécula | Nome da forma monomérica | Nome da forma polimérica | Exemplos de formas poliméricas |
|---|---|---|---|
| Aminoácidos | Aminoácidos | Proteínas (ou polipéptidos) | Proteínas fibrilares e proteínas globulares |
| Glícidos | Monossacarídeos | Polissacarídeos | Amido, glicogénio e celulose |
| Ácidos nucleicos | Nucleótidos | Polinucleótidos | ADN e ARN |
Aminoácidos e proteínas
As proteínas são compostas por aminoácidos dispostos numa cadeia linear e ligados entre si por ligações peptídicas. Muitas proteínas são as enzimas que catalisam as reacções químicas no metabolismo. Outras proteínas têm funções estruturais ou mecânicas, como o sistema de armação celular usado para manter a forma da célula, o citoesqueleto[5].
As proteínas desempenham também papéis importantes na sinalização celular, resposta imunitária, adesão celular, transporte activo através de membranas e no ciclo celular[6].
Lípidos
Os lípidos são o grupo mais diversificado de compostos bioquímicos. Constituem grande parte das membranas biológicas, tais como a membrana celular; além desta função estrutural, também servem como fonte de energia[6]. Os lípidos são normalmente definidos como moléculas biológicas hidrofóbicas ou anfipáticas solúveis em solventes orgânicos como o benzeno ou o clorofórmio[7].
As gorduras são um grupo alargado de compostos que inclui os ácidos gordos e o glicerol; uma molécula de glicerol ligada a três ácidos gordos por uma ligação éster é um triacilglicerol[8]. Existem diversas variações desta estrutura básica, incluindo a presença de esfingosina em esfingolípidos e grupos hidrofílicos como o fosfato nos fosfolípidos.
Os esteróides, como o colesterol, são outro grupo significativo de lípidos sintetizados em células.[9]
Glícidos

Os glícidos são aldeídos ou cetonas contendo diversos grupos funcionais hidroxilo. Os glícidos simples podem existir numa forma linear ou numa forma cíclica. São as moléculas biológicas mais abundantes e possuem funções muito diversificadas, como o armazenamento e transporte de energia (sob a forma de amido e glicogénio) e construção de elementos estruturais (como a celulose em plantas e a quitina em animais)[6].
Os glícidos mais simples são os monossacarídeos, que incluem a galactose, a frutose e a glicose. Os monossacarídeos podem formar polímeros designados polissacarídeos de formas muito diversas[10].
Nucleótidos
Os polímeros ADN e ARN são longas cadeias de nucleótidos. Estas macromoléculas são essenciais no armazenamento e uso da informação genética, através dos processos de transcrição e síntese proteica[6]. Esta informação é protegida por mecanismos de reparação do ADN e propagada através da replicação do ADN. Alguns vírus têm um genoma constituído por ARN (por exemplo, o HIV), que usam transcrição reversa para sintetizar ADN a partir desse ARN[11].
O ARN de ribozimas (como o spliceossoma) apresenta actividade enzimática tal como as enzimas proteicas, pois pode catalisar reacções químicas.
Os nucleósidos são sintetizados a partir da ligação de uma base azotada a uma ribose. Estas bases são anéis heterocíclicos contendo azoto, classificados como purinas ou pirimidinas. Os nucleótidos também actuam como coenzimas em reacções de transferência de grupos químicos[12].
Anabolismo e catabolismo
O metabolismo é normalmente dividido em dois grupo: anabolismo e catabolismo. Reacções anabólicas, ou reacções de síntese (produção de nova matéria), são reacções químicas que produzem nova matéria orgânica nos seres vivos. Sintetizam-se novos compostos (moléculas mais complexas) a partir de moléculas mais simples (com consumo de ATP). Reacções catabólicas, ou reacções de decomposição/degradação, são reacções químicas que produzem grandes quantidades de energia livre (sob a forma de ATP) a partir da decomposição ou degradação de moléculas mais complexas (matéria orgânica).
Quando o catabolismo supera em atividade o anabolismo, o organismo perde peso, o que acontece em períodos de jejum ou doença; mas se o anabolismo superar o catabolismo, o organismo cresce ou ganha peso. Se ambos os processos estão em equilíbrio, o organismo encontra-se em equilíbrio dinâmico ou homeostase.
Referências
- ↑ Friedrich C (1998). «Physiology and genetics of sulfur-oxidizing bacteria». Adv Microb Physiol. 39: 235-89. PMID 9328649
- ↑ Smith E, Morowitz H (2004). «Universality in intermediary metabolism». Proc Natl Acad Sci U S A. 101 (36): 13168-73. PMID 15340153
- ↑ Ebenhöh O, Heinrich R (2001). «Evolutionary optimization of metabolic pathways. Theoretical reconstruction of the stoichiometry of ATP and NADH producing systems». Bull Math Biol. 63 (1): 21–55. PMID 11146883
- ↑ Meléndez-Hevia E, Waddell T, Cascante M (1996). «The puzzle of the Krebs citric acid cycle: assembling the pieces of chemically feasible reactions, and opportunism in the design of metabolic pathways during evolution». J Mol Evol. 43 (3): 293–303. PMID 8703096
- ↑ Michie K, Löwe J (2006). «Dynamic filaments of the bacterial cytoskeleton». Annu Rev Biochem. 75: 467-92. PMID 16756499
- ↑ a b c d Nelson, David L.; Michael M. Cox (2005). Lehninger Principles of Biochemistry. New York: W. H. Freeman and company. 841 páginas. ISBN 0-7167-4339-6
- ↑ Fahy E, Subramaniam S, Brown H, Glass C, Merrill A, Murphy R, Raetz C, Russell D, Seyama Y, Shaw W, Shimizu T, Spener F, van Meer G, VanNieuwenhze M, White S, Witztum J, Dennis E (2005). «A comprehensive classification system for lipids». J Lipid Res. 46 (5): 839-61. PMID 15722563
- ↑ «Nomenclature of Lipids». IUPAC-IUB Commission on Biochemical Nomenclature (CBN). Consultado em 8 de março de 2007
- ↑ Hegardt F (1999). «Mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase: a control enzyme in ketogenesis». Biochem J. 338 (Pt 3): 569-82. PMID 10051425
- ↑ Raman R, Raguram S, Venkataraman G, Paulson J, Sasisekharan R (2005). «Glycomics: an integrated systems approach to structure-function relationships of glycans». Nat Methods. 2 (11): 817-24. PMID 16278650
- ↑ Sierra S, Kupfer B, Kaiser R (2005). «Basics of the virology of HIV-1 and its replication». J Clin Virol. 34 (4): 233-44. PMID 16198625
- ↑ Wimmer M, Rose I (1978). «Mechanisms of enzyme-catalyzed group transfer reactions». Annu Rev Biochem. 47: 1031–78. PMID 354490
Predefinição:Links Externos
Predefinição:Portal-bioquímica
Predefinição:Link FAPredefinição:Link FAPredefinição:Link FA
